Difference between revisions of "Diploma"
Marc Saric (talk | contribs) |
Marc Saric (talk | contribs) |
||
| (4 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | + | I did my biology diploma in biophysics, the title is: "Simulation von Large-Scale Motions bei der Substratbindung von Rubisco".<br /> It was part of a joint project of the [https://homepage.ruhr-uni-bochum.de/juergen.schlitter/ Theoretical Biophysics Group] and the [http://www.bpf.ruhr-uni-bochum.de Lehrstuhl für Biochemie der Pflanzen] . | |
| − | |||
| − | I did my biology diploma in biophysics, the title is: "Simulation von Large-Scale Motions bei der Substratbindung von Rubisco".<br /> It was part of a joint project of the [ | ||
---- | ---- | ||
| Line 7: | Line 5: | ||
== Molecular Modelling of Rubisco == | == Molecular Modelling of Rubisco == | ||
| − | [http:// | + | [http://xkcd.com/1039/ Ribulose-1,5-bisphosphate-Carboxylase/Oxygenase][http://en.wikipedia.org/wiki/RuBisCO (Rubisco for short)] is the most common protein in the biosphere of the earth. It´s purpose is to catalyze the addition of carbon dioxide on a small sugar-molecule (Ribulose-1,5-bisphosphate), the most important step in the dark- reaction of photosynthesis. |
My part of the work was the simulation and analysis of large conformational changes during the substrate-binding process with help of a newly developed GROMOS-based force-field (Schlitter et al.), which can simulate solvent-effects without simulating the solvent itself (thus leading to a tenfold increase in computation-speed compared to simulations with an explicit waterbox). | My part of the work was the simulation and analysis of large conformational changes during the substrate-binding process with help of a newly developed GROMOS-based force-field (Schlitter et al.), which can simulate solvent-effects without simulating the solvent itself (thus leading to a tenfold increase in computation-speed compared to simulations with an explicit waterbox). | ||
| Line 22: | Line 20: | ||
|[[Image:rubisco2.png|none|[[help:contents|Rubisco]]]] | |[[Image:rubisco2.png|none|[[help:contents|Rubisco]]]] | ||
|} | |} | ||
| − | |||
---- | ---- | ||
| − | If you are interested in some more in-depth-information about molecular modelling or Rubisco, this | + | If you are interested in some more in-depth-information about molecular modelling or Rubisco, this is definitely the wrong place, but you may look at the following papers, available from every university-library: |
G.F. Wildner et al.: Rubisco, an Old Challenge with new perspectives, Z. Naturforsch. 51 c, 263-276 (1996) | G.F. Wildner et al.: Rubisco, an Old Challenge with new perspectives, Z. Naturforsch. 51 c, 263-276 (1996) | ||
W.F. van Gunsteren & H.J.C. Berendsen: Moleküldynamik-Computersimulationen; Methodik, Anwendungen und Perspektiven in der Chemie, Angew.Chem. 102 (1990) 1020-1055 | W.F. van Gunsteren & H.J.C. Berendsen: Moleküldynamik-Computersimulationen; Methodik, Anwendungen und Perspektiven in der Chemie, Angew.Chem. 102 (1990) 1020-1055 | ||
Latest revision as of 01:12, 23 January 2021
I did my biology diploma in biophysics, the title is: "Simulation von Large-Scale Motions bei der Substratbindung von Rubisco".
It was part of a joint project of the Theoretical Biophysics Group and the Lehrstuhl für Biochemie der Pflanzen .
Molecular Modelling of Rubisco
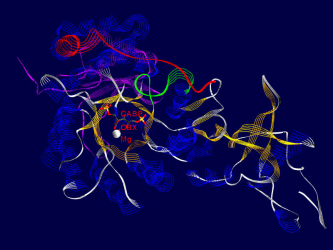
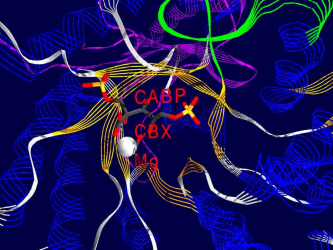
Ribulose-1,5-bisphosphate-Carboxylase/Oxygenase(Rubisco for short) is the most common protein in the biosphere of the earth. It´s purpose is to catalyze the addition of carbon dioxide on a small sugar-molecule (Ribulose-1,5-bisphosphate), the most important step in the dark- reaction of photosynthesis.
My part of the work was the simulation and analysis of large conformational changes during the substrate-binding process with help of a newly developed GROMOS-based force-field (Schlitter et al.), which can simulate solvent-effects without simulating the solvent itself (thus leading to a tenfold increase in computation-speed compared to simulations with an explicit waterbox).
Pictures
One of the fascinating parts of computer simulation of molecular processes is the fact that you get incredibly nice graphics ;-)
Here are some examples of what Rubisco looks like, together with some short interpretation of the structure and function of the molecule.
All pictures are made from the 8RUB.PDB-structure from the Brookhaven Protein-Database (afaik this structure has been replaced by a better one recently), and have been rendered using SwissPDBViewer/DeepView and Povray , both programs based on WIN95 running on my P133, 24 MB back in 1999.
If you are interested in some more in-depth-information about molecular modelling or Rubisco, this is definitely the wrong place, but you may look at the following papers, available from every university-library:
G.F. Wildner et al.: Rubisco, an Old Challenge with new perspectives, Z. Naturforsch. 51 c, 263-276 (1996)
W.F. van Gunsteren & H.J.C. Berendsen: Moleküldynamik-Computersimulationen; Methodik, Anwendungen und Perspektiven in der Chemie, Angew.Chem. 102 (1990) 1020-1055